Research Activity
Most real-life applications of statistics require the use of computational methods.
This is particularly true for Bayesian statistics, where the output of an analysis is the posterior distribution — a distribution over models and parameters that quantifies the uncertainty of inferences from the data. In almost all applications the posterior distribution is intractable, and instead of analytical evaluation of probabilities and expectations we use algorithms to draw samples from the posterior. These algorithms are important in many areas of statistics, particularly when we need to average over uncertainty within our statistical models.
At Lancaster we have expertise in a range of Bayesian computational methods, including Markov chain Monte Carlo, sequential Monte Carlo and approximate Bayesian computation. A particular focus of the group is on developing new algorithms that have excellent computational properties in settings where we wish to fit complex stochastic models to large datasets. Our research is collaborative, with involvement in large multi-institutional projects that are aiming to develop the next generation of methods, motivated by applications ranging from the Health Sciences, to Engineering and Security.
The research group meets weekly over coffee, where we discuss recent papers, present our own new research ideas and hear directly from external researchers on their latest innovations. Past talks and discussions are detailed on Github.
Case Study: The Apogee to Apogee Path Sampler
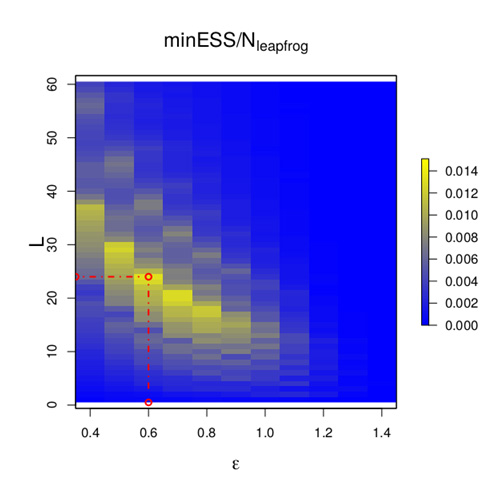
Hamiltonian Monte Carlo
Hamiltonian Monte Carlo (HMC) is, perhaps, the most commonly used Markov chain Monte Carlo technique in the world; however, it is notoriously difficult to tune. A transformation of the Bayesian posterior can be viewed as a very high-dimensional, irregular U-shaped surface, and at the start of each iteration of the HMC algorithm, the current value can be viewed as a small ball on this surface. The algorithm then “kicks” the ball in a random direction and follows it by numerically solving the equations of motion using a time-discretisation, epsilon. The ball’s position after some time, T=L x epsilon, is the potential next point of the algorithm, and should, ideally, be a good distance from the current point. This suggests choosing a large value for L; however, if L is too large then the ball will roll up the side of the U and back down, perhaps ending its journey very close to where it started. This shows that epsilon and L must be “just right” for HMC to achieve close to its optimal efficiency.
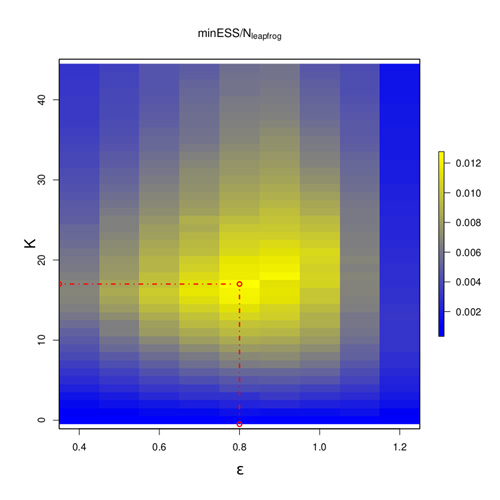
Apogee to Apogee Path Sampler
The Apogee to Apogee Path Sampler (AAPS) relies on the same transformation of the surface as HMC but looks backwards in time as well as forwards and splits the path of the ball into segments according to the locally highest points, or apogees, reached along its trajectory around the multi-dimensional U. It also chooses the potential next point randomly from the whole trajectory, with a bias towards points further from the start, rather than forcing it to be the very end point. The AAPS achieves similar efficiencies to those of HMC; however, as can be seen from this figure, its efficiency is very insensitive to the choice of its tuning parameters (epsilon and the number of apogees, K) compared to HMC.
-
Group Members
Loading People
Projects
DSI: COVID-19: Bayesian inference for high resolution stochastic modelling for the UK
01/08/2021 → 31/01/2023
Research
DSI: A critical assessment of mobility models to quantify infections and interventions
01/04/2021 → 31/03/2026
Research
COVID-19 Modelling Consortium: Quantitative epidemiological predictions in response to an evolving pandemic.
19/11/2020 → 31/03/2023
Research
DSI: Klebsiella transmission and immunity on Chatinkha nursery
01/11/2020 → 30/09/2023
Research
DSI: Covid: SARS-CoV-2 immunoepidemiology in Wellcome-funded urban and rural cohorts in Malawi: generating evidence to inform regional medium and long term decision making
01/09/2020 → 28/02/2022
Research
National COVID-19 Wastewater Epidemiology Surveillance Programme
06/07/2020 → 05/11/2021
Research
Tensorflow Probability
02/03/2020 → …
Research
Elimination of endemic livestock disease in low and middle income countries
01/12/2019 → 31/01/2026
Research
DSI: GEM: translational software for outbreak analysis
01/11/2019 → 01/05/2021
Research
DSI: GEM: translational software for outbreak analysis
01/11/2019 → 01/05/2021
Research
SAVSNet Agile: Responsible data intelligence for canine health
01/08/2019 → 31/07/2022
Research
DSI: CoSInES: COmputational Statistical INference for Engineering and Security
01/10/2018 → 30/09/2024
Research
Drivers of resistance in Uganda and Malawi: The DRUM Consortium
01/05/2018 → 31/10/2021
Research
New Approaches to Bayesian Data Science: Tackling Challenges from the Health Sciences
01/04/2018 → …
Research
New Approaches to Bayesian Data Science: Tackling Challenges from the Health Sciences
01/04/2018 → 31/07/2024
Research
Quantitative analysis and modelling on targets and strategies for alleviating the burden of priority neglected tropical diseases
01/03/2018 → 28/02/2021
Research
Spatiotemporal modelling for bovine tuberculosis associations in badger and cattle populations in England
01/09/2017 → 31/05/2019
Research
DSI:LHOFT - Liverpool-Humber Optimisation of Freight Transport
01/08/2017 → 31/01/2021
Research
Farming Food and Forecasting
01/04/2017 → 31/03/2022
Research




















