| o
Tomato 14-3-3 genes |
|
o
oDefence response target proteins |
| Tobacco 14-3-3 genes | oAntisense 14-3-3 plants | |
| o | o |
| o
Tomato 14-3-3 genes |

|
o
oDefence response target proteins |
| Tobacco 14-3-3 genes | oAntisense 14-3-3 plants | |
| o | o |
|
|
||
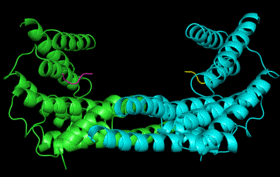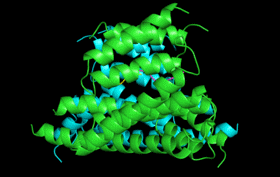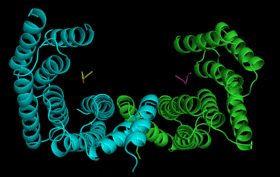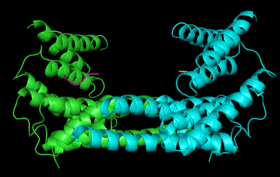
| 14-3-3 Proteins | Plant defence signalling | 14-3-3 structure/function |
14-3-3
proteins are found in all eukaryotes where they function as regulators
of a wide range of biological processes. They function by interacting directly
with numerous different target proteins thereby altering their activity.
Interactions are generally mediated by phosphorylation of specific binding
sites in the target proteins. 14-3-3 binding can either:
|
|
As
well as taking molecular genetic and biochemical approaches to investigate
14-3-3 protein-protein interactions in plant defence responses, we are
also interested to determine whether and how different 14-3-3 isoforms
play specific roles in the plant. This will be achieved through:
|